If you find Complexity Thoughts, click on the Like button, leave a comment, repost on Substack or share this post. It is the only feedback I can have for this free service.
The frequency and quality of this newsletter relies on social interactions. Thank you!
Dall-e 3 representation of this issue’s content
In a nutshell
We have again works from a variety of disciplines, including foundational/theoretical studies and computational/experimental ones.
Writing this summary is becoming increasingly difficult with such a variety of topics, therefore I am going to test a new approach: besides reporting the abstract, and accompanying it with a specific personal comment, I will also highlight a specific sentence that attracted my attention in the paper.
What do you think? Can it work better than attempting to write a consistent summary of so many different studies in a paragraph? Let me know through the comments and enjoy Issue #21.
Foundations of network science and complex systems
Community Detection with the Map Equation and Infomap: Theory and Applications
A great review on finding the mesoscale organization of complex networks by exploiting how information flows through them:
Real-world networks have a complex topology comprising many elements often structured into communities. Revealing these communities helps researchers uncover the organizational and functional structure of the system that the network represents. However, detecting community structures in complex networks requires selecting a community detection method among a multitude of alternatives with different network representations, community interpretations, and underlying mechanisms. This review and tutorial focuses on a popular community detection method called the map equation and its search algorithm Infomap. The map equation framework for community detection describes communities by analyzing dynamic processes on the network. Thanks to its flexibility, the map equation provides extensions that can incorporate various assumptions about network structure and dynamics. To help decide if the map equation is a suitable community detection method for a given complex system and problem at hand -- and which variant to choose -- we review the map equation's theoretical framework and guide users in applying the map equation to various research problems.
Our results thus highlight the need for good reconstructions of low-order interactions in order to have reliable reconstructions of full hypergraphs
Complex networked systems are influenced by interactions between multiple elements, known as hyperedges. Researchers have used inference methodologies to understand hypergraph structure, assuming that the statistical mechanism behind hyperedges is assortative, and relying on a specific set of groups to explain interactions. However, our results show that these assumptions are not always accurate. The same groups of nodes can explain hyperedges of different sizes, but interactions between nodes are not always assortative. Nonassortative patterns can also explain some observed hyperedges. This research challenges existing inference methodologies and broadens understanding of complex networked systems, encouraging the development of new approaches to studying hypergraphs.
Geometric description of clustering in directed networks
First-principle network models are crucial to understanding the intricate topology of real complex networks. Although modelling efforts have been quite successful in undirected networks, generative models for networks with asymmetric interactions are still not well developed and unable to reproduce several basic topological properties. Progress in this direction is of particular interest, as real directed networks are the norm rather than the exception in many natural and human-made complex systems. Here we show how the network geometry paradigm can be extended to the case of directed networks. We define a maximum entropy ensemble of random geometric directed graphs with a given sequence of in-degrees and out-degrees. Beyond these local properties, the ensemble requires only two additional parameters to fix the levels of reciprocity and the frequency of the seven possible types of three-node cycles in directed networks. A systematic comparison with several representative empirical datasets shows that fixing the level of reciprocity alongside the coupling with an underlying geometry is able to reproduce the wide diversity of clustering patterns observed in real directed complex networks.
Human Behavior
The effect of natural disasters on human capital in the United States
The authors quantify an important loss of human capital due to natural disasters in the U.S. We need more studies like this to go beyond purely structural damage assessment of climate change:
Although natural disasters are commonplace, they leave in their wake an enormous amount of damage. The physical damage they cause is immediately apparent, but less obvious is the potential magnitude of disruptions to learning and resulting damage to human capital. Using the universe of Presidential Disaster Declarations in the United States, we show that natural disasters impact a region’s human capital both via reductions in learning for students who remain in school as well as a reduction in the years of schooling completed. These effects appear to be scarring and persistent. Quantifying these losses using the implied reduction of lifetime earnings suggests that natural disasters reduce a region’s human capital by a similar magnitude as the assessed property damage.
The unequal effects of the health–economy trade-off during the COVID-19 pandemic
This study provides a representative approach to attack complex problems involving the interplay of complex systems. Likely, it will trigger the research needed to cope with the next pandemic:
Despite the global impact of the coronavirus disease 2019 pandemic, the question of whether mandated interventions have similar economic and public health effects as spontaneous behavioural change remains unresolved. Addressing this question, and understanding differential effects across socioeconomic groups, requires building quantitative and fine-grained mechanistic models. Here we introduce a data-driven, granular, agent-based model that simulates epidemic and economic outcomes across industries, occupations and income levels. We validate the model by reproducing key outcomes of the first wave of coronavirus disease 2019 in the New York metropolitan area. The key mechanism coupling the epidemic and economic modules is the reduction in consumption due to fear of infection. In counterfactual experiments, we show that a similar trade-off between epidemic and economic outcomes exists both when individuals change their behaviour due to fear of infection and when non-pharmaceutical interventions are imposed. Low-income workers, who perform in-person occupations in customer-facing industries, face the strongest trade-off.
(Network) Neuroscience
Functional neuroimaging as a catalyst for integrated neuroscience
Interesting Perspective piece on the potential role of fMRI to integrate a variety of subfields in neuroscience:
Functional magnetic resonance imaging (fMRI) enables non-invasive access to the awake, behaving human brain. By tracking whole-brain signals across a diverse range of cognitive and behavioural states or mapping differences associated with specific traits or clinical conditions, fMRI has advanced our understanding of brain function and its links to both normal and atypical behaviour. Despite this headway, progress in human cognitive neuroscience that uses fMRI has been relatively isolated from rapid advances in other subdomains of neuroscience, which themselves are also somewhat siloed from one another. In this Perspective, we argue that fMRI is well-placed to integrate the diverse subfields of systems, cognitive, computational and clinical neuroscience. We first summarize the strengths and weaknesses of fMRI as an imaging tool, then highlight examples of studies that have successfully used fMRI in each subdomain of neuroscience. We then provide a roadmap for the future advances that will be needed to realize this integrative vision. In this way, we hope to demonstrate how fMRI can help usher in a new era of interdisciplinary coherence in neuroscience.
Learning how network structure shapes decision-making for bio-inspired computing
To better understand how network structure shapes intelligent behavior, we developed a learning algorithm that we used to build personalized brain network models for 650 Human Connectome Project participants. We found that participants with higher intelligence scores took more time to solve difficult problems, and that slower solvers had higher average functional connectivity. With simulations we identified a mechanistic link between functional connectivity, intelligence, processing speed and brain synchrony for trading accuracy with speed in dependence of excitation-inhibition balance. Reduced synchrony led decision-making circuits to quickly jump to conclusions, while higher synchrony allowed for better integration of evidence and more robust working memory. Strict tests were applied to ensure reproducibility and generality of the obtained results. Here, we identify links between brain structure and function that enable to learn connectome topology from noninvasive recordings and map it to inter-individual differences in behavior, suggesting broad utility for research and clinical applications.
Biological Systems
Seeing around corners: Cells solve mazes and respond at a distance using attractant breakdown
I think that this summarizes it all:
In many cases, self-generated gradients allow cells to steer with an accuracy that seems intuitively impossible
Self-generated chemoattractant gradients allow cells to navigate complex paths with great efficiency. Diffusion and attractant breakdown allow cells to obtain detailed information about their surroundings that could not be provided by simple attractant gradients. Cell migration in vivo cannot be understood without considering the interplay among cells, attractants, and the structure of the local environment. Microfluidic mazes are an excellent experimental system for studying these interactions
Future medicine: from molecular pathways to the collective intelligence of the body
There is a lot of food for thoughts in this Review, but I especially like how complexity science is heavily used to tackle current challenges in medicine (eg, inducing organ repair) and the focus on collective computation to health from a systemic perspective.
It is likely that the properties of brains that enable control via highly efficient high-level interactions are not unique – and are instead accelerated and modified versions of universal cellular capabilities that evolution pivots into diverse problem spaces as needed
Recent advances in research on diverse kinds of intelligence in non-neural and other unconventional substrates leverage insights from cognitive neuroscience and collective computation to offer a new approach to system-level health: communication and behavior-shaping of the activity of cellular swarms.
Regenerative medicine can exploit the native problem-solving of molecular and cellular networks to induce complex organ repair and cancer reprogramming.
Bioelectric networks formed by somatic cells offer a highly tractable interface to exploit the collective intelligence of cells and tissues. Preclinical studies, many with FDA-approved drugs, demonstrate applications for birth defects, appendage regeneration, and cancer suppression.
Implanted hepatocytes build an ectopic liver exactly tuned to replace lost function, offering the prospect of clinical application for end-stage organ failure, and a vision for further therapies based on inherent cell capabilities.
The computational capabilities of many-to-many protein interaction networks
Many biological circuits comprise sets of protein variants that interact with one another in a many-to-many, or promiscuous, fashion. These architectures can provide powerful computational capabilities that are especially critical in multicellular organisms. Understanding the principles of biochemical computations in these circuits could allow more precise control of cellular behaviors. However, these systems are inherently difficult to analyze, due to their large number of interacting molecular components, partial redundancies, and cell context dependence. Here, we discuss recent experimental and theoretical advances that are beginning to reveal how promiscuous circuits compute, what roles those computations play in natural biological contexts, and how promiscuous architectures can be applied for the design of synthetic multicellular behaviors.
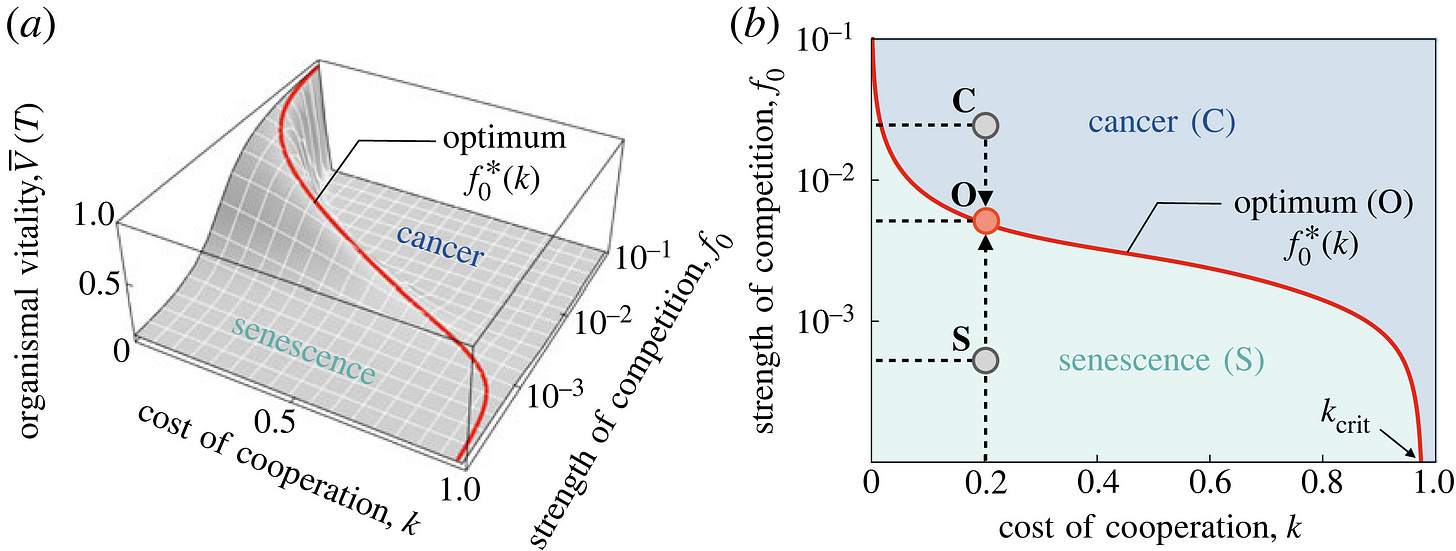
Optimal intercellular competition in senescence and cancer
Another great complex systems approach to understanding the emergence of multicellularity:
an optimal level of intercellular competition in a multicellular organism maximizes organismal vitality by delaying the inevitability of ageing
Effective multicellularity requires both cooperation and competition between constituent cells. Cooperation involves sacrificing individual fitness in favour of that of the community, but excessive cooperation makes the community susceptible to senescence and ageing. Competition eliminates unfit senescent cells via natural selection and thus slows down ageing, but excessive competition makes the community susceptible to cheaters, as exemplified by cancer and cancer-like phenomena. These observations suggest that an optimal level of intercellular competition in a multicellular organism maximizes organismal vitality by delaying the inevitability of ageing. We quantify this idea using a statistical mechanical framework that leads to a generalized replicator dynamical system for the population of cells that changes their vitality and cooperation due to somatic mutations that make them susceptible to ageing and/or cancer. By accounting for the cost of cooperation and strength of competition in a minimal setting, we show that our model predicts an optimal value of competition that maximizes vitality and delays the inevitability of senescence or cancer. The results have implications for the design of strategies aimed at delaying ageing in biological, technical and social systems that exhibit similar processes.
From microbiome composition to functional engineering, one step at a time
As a non-expert on microbiome, I think that this Review does a great job, especially in highlighting the current knowledge gaps about interactions:
Communities of microorganisms (microbiota) are present in all habitats on Earth and are relevant for agriculture, health, and climate. Deciphering the mechanisms that determine microbiota dynamics and functioning within the context of their respective environments or hosts (the microbiomes) is crucially important. However, the sheer taxonomic, metabolic, functional, and spatial complexity of most microbiomes poses substantial challenges to advancing our knowledge of these mechanisms. While nucleic acid sequencing technologies can chart microbiota composition with high precision, we mostly lack information about the functional roles and interactions of each strain present in a given microbiome. This limits our ability to predict microbiome function in natural habitats and, in the case of dysfunction or dysbiosis, to redirect microbiomes onto stable paths. Here, we will discuss a systematic approach (dubbed the N+1/N−1 concept) to enable step-by-step dissection of microbiome assembly and functioning, as well as intervention procedures to introduce or eliminate one particular microbial strain at a time. The N+1/N−1 concept is informed by natural invasion events and selects culturable, genetically accessible microbes with well-annotated genomes to chart their proliferation or decline within defined synthetic and/or complex natural microbiota. This approach enables harnessing classical microbiological and diversity approaches, as well as omics tools and mathematical modeling to decipher the mechanisms underlying N+1/N−1 microbiota outcomes. Application of this concept further provides stepping stones and benchmarks for microbiome structure and function analyses and more complex microbiome intervention strategies.
The molecular basis for cellular function of intrinsically disordered protein regions
A must read if you are looking for an updated Review about disordered proteins, which are crucial a variety of cellular processes:
Intrinsically disordered protein regions exist in a collection of dynamic interconverting conformations that lack a stable 3D structure. These regions are structurally heterogeneous, ubiquitous and found across all kingdoms of life. Despite the absence of a defined 3D structure, disordered regions are essential for cellular processes ranging from transcriptional control and cell signalling to subcellular organization. Through their conformational malleability and adaptability, disordered regions extend the repertoire of macromolecular interactions and are readily tunable by their structural and chemical context, making them ideal responders to regulatory cues. Recent work has led to major advances in understanding the link between protein sequence and conformational behaviour in disordered regions, yet the link between sequence and molecular function is less well defined. Here we consider the biochemical and biophysical foundations that underlie how and why disordered regions can engage in productive cellular functions, provide examples of emerging concepts and discuss how protein disorder contributes to intracellular information processing and regulation of cellular function.
Oldies but Goodies
The bacterium as a model neuron
At first glance, a unicellular organism such as a bacterium would seem to be an unlikely model for a neural cell, which in contrast forms part of an intricate network in a multicellular organism. Indeed, if one is seeking to learn the wiring diagram of a complex neural system, then the study of bacterial behavior has little to offer. If, on the other hand, one is seeking to learn how signals are processed within a cell or how this processing machinery can be modified to produce plasticity, then there appear to be many points of analogy. In this article we shall consider the properties of the bacterial cell, its receptors, memory system, and output response, emphasizing those attributes which may be relevant to information processing in higher species.
Intracellular signalling as a parallel distributed process
Living cells respond to their environment by means of an interconnected network of receptors, second messengers, protein kinases and other signalling molecules. This article suggests that the performance of cell signalling pathways taken as a whole has similarities to that of the parallel distributed process networks (PDP networks) used in computer-based pattern recognition. Using the response of hepatocytes to glucagon as an example, a procedure is described by which a PDP network could simulate a cell signalling pathway. This procedure involves the following steps: (a) a bounded set of molecules is defined that carry the signals of interest; (b) each of these molecules is represented by a PDP-type of unit, with input and output functions and connection weights corresponding to specific biochemical parameters; (c) a “learning algorithm” is applied in which small random changes are made in the parameters of the cell signalling units and the new network is then tested by a selection procedure in favour of a specific input-output relationship. The analogy with PDP networks shows how living cells can recognize combinations of environmental influences, how cell responses can be stabilized and made resistant to damage, and how novel cell signalling pathways might appear during evolution.











So many exciting papers!
Thank you for sharing! enrich my days!